- >>
- Healthy Adult Studies
- HCP Young Adult
- Project Protocols Detail
Logistics of Data Acquisition
We plan a multi-track data acquisition strategy for optimal utilization of the equipment and the large subject population available to our consortium.
During the optimization phase (Phase I, Fall 2010 - Spring 2012), pilot data is being acquired at four institutions: 3T data at Washington University and at UC Berkeley, 3T and 7T data at University of Minnesota, and MEG/EEG data at St. Louis University.
During Phase II (Summer 2012 - Summer 2015), all 1,200 subjects in the main cohort will be scanned at Washington University on a dedicated 3T scanner optimized for HARDI (High Angular Resolution Diffusion Imaging), R-fMRI (Resting-state fMRI), and T-fMRI (task-evoked fMRI). A subset of 200 subjects (100 same-sex twin pairs, 50% MZ) will also be scanned at University of Minnesota using 7T MRI (HARDI, R-fMRI, and T-fMRI). Another subset of same-sex monozygotic twin pairs will be scanned at St. Louis University using MEG/EEG (both resting and task-evoked).
The dedicated 3T scanner at WashU (Washington University) will be a customized device with about double the standard gradient strength. This will provide high-throughput capability, high reliability, and geographic proximity to the sibship cohort. It will yield an excellent reference dataset for future connectome studies carried out at various institutions that have a 3T scanner but lack a 7T capability.
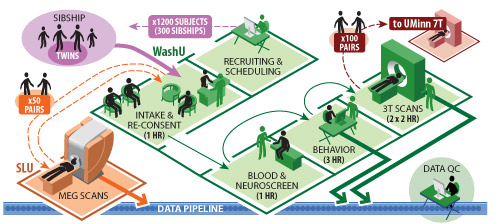
Figure 1: Overview of the logistics of data acquisition at three main sites.
7T scanning at UMinn (University of Minnesota) will leverage the extensive experience of the UMinn group that pioneered 7T human imaging. Having the 7T scans done on individuals also scanned at 3T will allow higher-resolution 7T data to constrain and better interpret the 3T data.
For Phase II, high-throughput, production-mode operations will enable data acquisition from WashU, UMinn, and SLU. Fig. 1 shows the overall workflow during Phase II for components related to subject recruitment and scheduling of all subjects by the WashU team followed by data acquisition at each of the three sites.