- >>
- Healthy Adult Studies
- HCP Young Adult
- News
- Article
Release of S1200 Extensively Processed rfMRI data
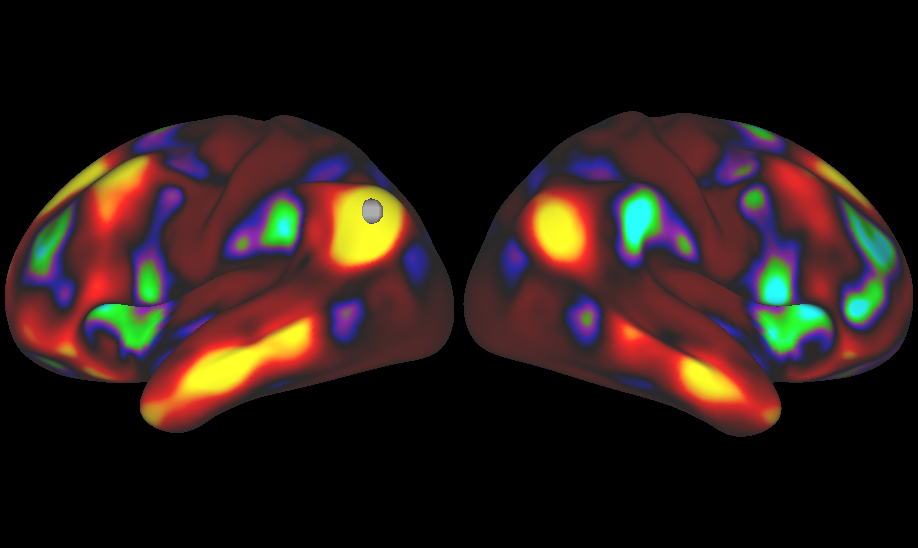 The Human Connectome Project (HCP) WU-Minn consortium is pleased to announce the release of extensively processed 3T rfMRI data associated with the HCP S1200 Data Release, the final release of subjects from the Young Adult HCP. This release includes group average dense functional connectomes and the
“PTN” (Parcellation + Timeseries + Netmats) dataset that can be downloaded from the S1200 Project page in ConnectomeDB. Upon login, click "Open Dataset" or use this link: https://db.humanconnectome.org/data/projects/HCP_1200.
The Human Connectome Project (HCP) WU-Minn consortium is pleased to announce the release of extensively processed 3T rfMRI data associated with the HCP S1200 Data Release, the final release of subjects from the Young Adult HCP. This release includes group average dense functional connectomes and the
“PTN” (Parcellation + Timeseries + Netmats) dataset that can be downloaded from the S1200 Project page in ConnectomeDB. Upon login, click "Open Dataset" or use this link: https://db.humanconnectome.org/data/projects/HCP_1200.
These analyses are based on data from all 1003 subjects (fMRI recon r177 + r227) in the S1200 data release having four complete 3T rfMRI runs (4800 timepoints collected), or a subset of 812 of these subjects whose fMRI data was reconstructed using the improved r227 recon algorithm that was in place in the latter two thirds of the HCP project.
Complete dataset information, including an Appendix which shows a comparison of the 1003 all subjects data and the 812 "recon 2" data, is provided in the documentation for this release: http://humanconnectome.org/storage/app/media/documentation/s1200/HCP1200-DenseConnectome+PTN+Appendix-July2017.pdf
HCP_S1200 group average functional connectivity. MSM-All registered group average dense (grayordinatesXgrayordinates) functional connectomes have been generated from group-PCA outputs for each of the S1200 subjectsgroups: 1003 subjects (recon version r177 + r227) and 812 subjects (r227 only). These large (33 GB) files may be downloaded from the S1200 Project page directly, generated from the group-PCA Eigenmaps also available using scripts provided in the PTN dataset, or can be accessed remotely in Connectome Workbench wb_view (recommended) through links as explained in the release document.
PTN (Parcellation + Timeseries + Netmats) datasets. Group-PCA outputs were further processed each of the S1200 subjects groups described above ,yielding the following outputs:
1. Group-average “parcellations”, obtained by means of group-ICA at several different dimensionalities (15, 25, 50, 100, 200, 300).
2. Subject-specific sets of “node timeseries” – for each subject, a representative time series per ICA component (“parcel”).
3. A subject-specific “parcellated connectome” – for each subject, a nodes x nodes matrix – the functional connectivity between node timeseries (available for the 1003 subject group analysis only as separate downloads for various ICA dimensionalities).
Stay tuned for the forthcoming release of other S1200 group average data, including group average surfaces and volumes, and a scene file and tutorial to facilitate viewing these datasets in Connectome Workbench wb_view!