- >>
- Healthy Adult Studies
- HCP Young Adult
- Documentation Detail
Creating and Using Subject Keys in ConnectomeDB
In any paper that links individual subject data and restricted data for subjects (e.g. twin status, family structure, exact age, BMI, etc.) from the HCP dataset, authors must generate their own Coded IDs rather than use the original HCP subject ID. In ConnectomeDB, we allow authors to upload a Subject Key: a collection of Coded IDs that link to the HCP IDs for each cited subject.
Restricted Access Required! Only HCP users with restricted data access may create or view these Subject Keys.
How to Create a Subject Key
Follow these step by step instructions:
Naming your Subject Key
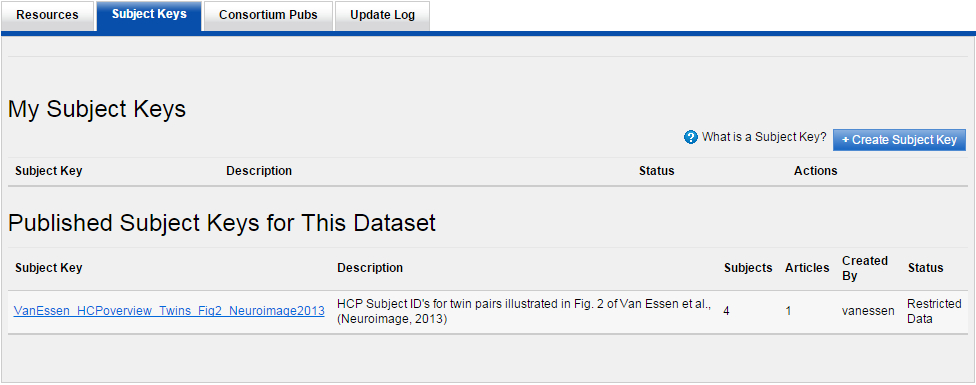
1. Log in to ConnectomeDB and browse to the WU-Minn HCP Data - 1200 Subjects section and click "Open Dataset".
2. Click on the "Subject Keys" tab halfway down the page. Click the button that says "Create Subject Key". If necessary, switch into Restricted Data viewing mode to continue.
3. Create a title and description for your subject key. This metadata can be seen by all ConnectomeDB users with project access, so please refrain from including restricted or identifying information in your title.
Entering your Coded IDs
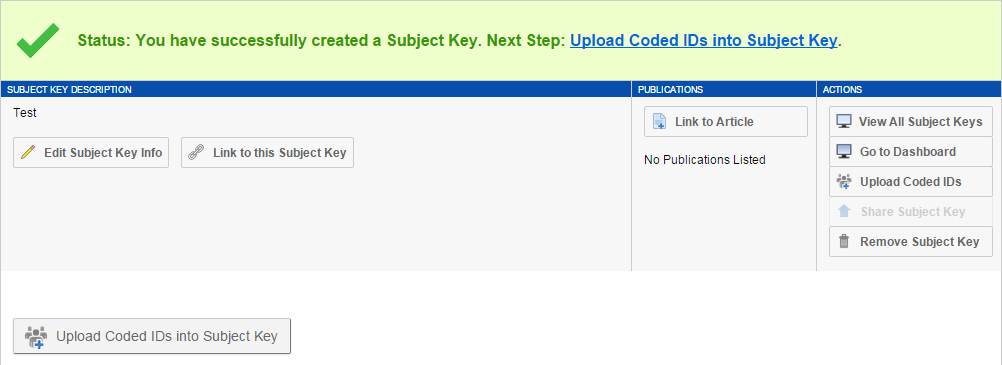
4. Click the "Upload Coded IDs Into Subject Key" button on the Subject Key report page.
5. Create a simple spreadsheet associating your Coded IDs with the original HCP Subject IDs, and save it as a CSV file. We recommend adding your Subject IDs and Coded IDs to the template file available to download.
6. Upload your spreadsheet. Any errors found in your file will be identified and reported to you before your data is saved.
7. Click "Save your Coded IDs to your Subject Key"
Warning for first-time users! Due to a known issue in ConnectomeDB, the first time that you attempt to upload a CSV file to your Subject Key, you may receive an error screen that incorrectly states that you do not have permission to continue. If this happens, navigate back to the home page, log out, then log back in. You will be able to find your Subject Key on the dataset home page and continue the process from Step 8.
Add Publication info
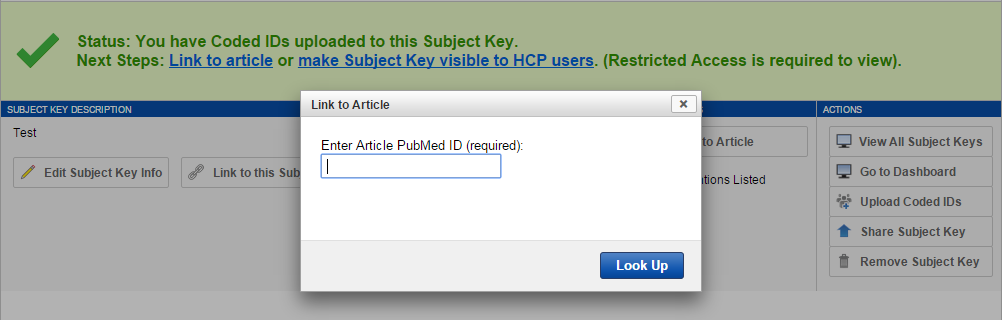
8. Click the "Add Article" button on the Subject Key report page.
9. Enter your article's PMID in the PubMed search field. The PMID is a string of digits used to uniquely identify your article. The easiest place to find it is in the URL of your article's PubMed entry. Example: http://www.ncbi.nlm.nih.gov/pubmed/23684880.
10. Click "Look Up" to query PubMed and retrieve your article's metadata. This will be automatically saved to your Subject Key. (You can remove articles at any time by clicking the "Remove" icon in the list of articles.)
Publish your Subject Key
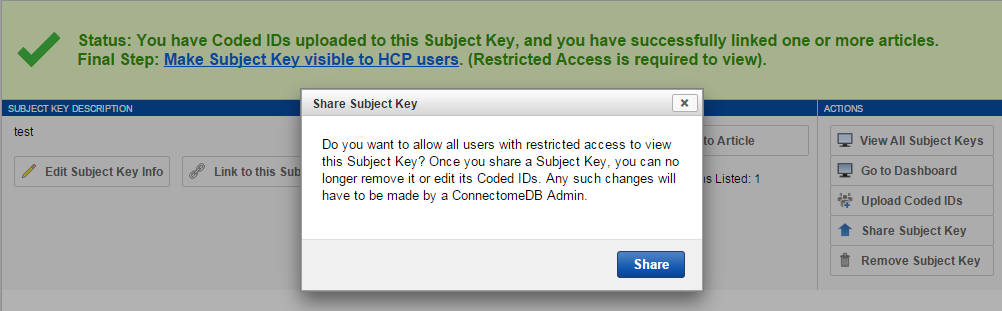
11. Click "Share Subject Key". With Coded IDs added and articles associated, you can now publish your Subject Key for other users to see.