- >>
- Healthy Adult Studies
- HCP Young Adult
- News
- Article
Workbench beta version 0.81 released
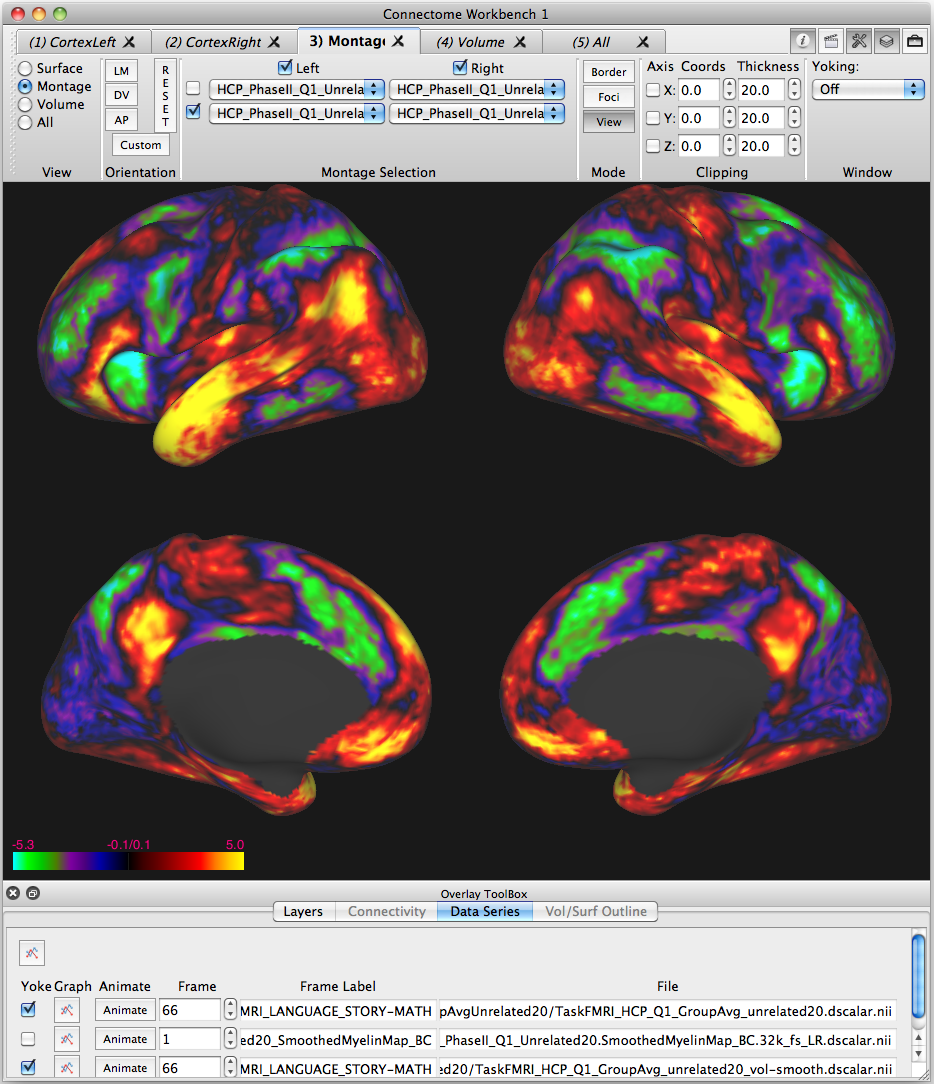
HCP has released the newest beta version of its Connectome Workbench brain surface/volume visualization and analysis software to coincide with the HCP Quarter 1 data release. Workbench software is freely available to all on the HCP website.
Several features have been updated in v0.81 beta, including a redesigned interface to open files remotely (e.g. large connectivity files from ConnectomeDB), more complete scene file saving capabilities, improvements to map thresholding and border/foci drawing, and the addition of various Workbench command operations.
If you are not yet familiar with Workbench, here are a few basic questions and answers to get you up to speed:
Why use Connectome Workbench?
Workbench software is being specifically designed for interactive viewing of data associated with the HCP, especially multi-dimensional time series, scalar, and connectivity maps in the relatively new CIFTI format that incorporates surface vertices (both hemispheres) plus subcortical gray-matter voxels in a single file format. While you can view HCP files that are in standard NIFTI and GIFTI format using other brain-mapping software platforms, currently only Connectome Workbench is compatible with CIFTI formatted data.
If you give it a try, you will also see that we have put a considerable effort into making Workbench easy to use, even for beginners, while it still serves the purposes of more advanced users.
How do I learn to use Workbench?
Tutorials and associated datasets are available to help you quickly explore using Workbench, or take a deeper dive into its detailed functionality. The Q1 Workbench Tutorial is written for taking a short tour of the additionally processed group average data (myelin maps, task fMRI contrasts, and functional connectivity) for 20 Q1 participants. The Beta v0.7 Workbench Tutorial offers a more comprehensive guide, beginning with a chapter that shows you how to navigate through several types of brain mapping data, then providing more detail in subsequent chapters, including instructions on how to import your own data into Workbench.
Why the “beta”?
Although earlier versions of Workbench have been publicly available since last June, the software is still considered “beta” because a considerable amount of functionality for data being generated by HCP analysis pipelines (which are themselves under active development) has yet to be included in the software.
What can I look forward to in future versions of Workbench?
Some features on the horizon are: capabilities for interactive viewing of probabilistic tractography, viewing of parcellated functional connectivity, ability to create custom color palettes, improved graphing of data series (eg. resting state timeseries or ICA components graphed by vertex), and more seamless interactivity with ConnectomeDB.