- >>
- Healthy Adult Studies
- HCP Young Adult
- Project Protocols Detail
Components of the Human Connectome Project - MEG and EEG
MEG Data Released:
High-quality data for 67 subjects has been released on ConnectomeDB.
Learn more
MEG (magnetoencephalography) and EEG (electroencephalography) are non-invasive electrophysiological techniques for recording brain activity. EEG is based on measuring voltage sensed by an array of electrodes placed on the scalp. MEG is based on measuring the magnetic field outside the head using an array of very sensitive magnetic field detectors (magnetometers). The signals recorded by EEG and MEG directly reflect current flows generated by neurons within the brain. The temporal frequency content of these signals ranges from less than 1 Hz (one cycle per second) to over 100 Hz (100 cycles per second). This rich spectral content offers a view of the brain operating in "real time."
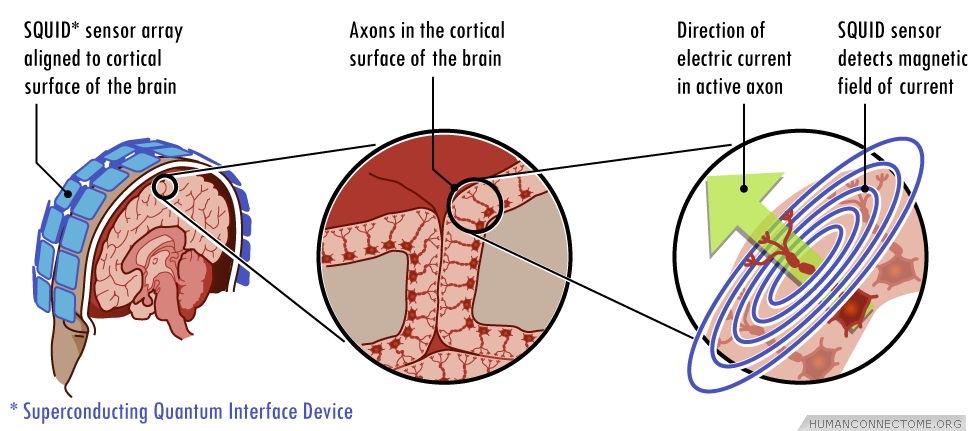
Signals such as those illustrated in the plot below are recorded from each MEG sensor or EEG electrode pair. From MEG/EEG signals its easy to build a surface map reflecting ongoing brain activity, however, building 3D maps of brain activity from these signals requires the solution of an inverse source localization problem that finds the locations in the brain of sources of electromagnetic activity. The inverse source localization of MEG/EEG signals is accomplished by selecting a suitable model for brain activity such as the current dipole model. Within this framework, the best dipole distribution accounting for the observed magnetic field and/or electric potential is found. The resolution of MEG/EEG strongly relies on the accuracy of the chosen model and is usually of the order of ~1–2 cm. Moreover, the accuracy of the inverse source localization of MEG generators degrades in subcortical regions. Thus, often only the cortex is modeled in MEG/EEG studies.
Once a source spatial activation map is generated, it is superimposed onto a 3-D representation of the subjects brain anatomy derived from high resolution MRI data. Thus, electrophysiological activity sources within the brain are typically shown highlighted or colored very much as are representations of fMRI responses (see Resting-State MEG). The limited localization accuracy and inherent spatial resolution of MEG/EEG means that the resulting activation maps are more diffuse and less anatomically precise than corresponding fMRI derived maps, but, they can be represented as movies to show how activation patterns change in real time as the brain functions.
Figure 2: An example of MEG/EEG time signals.
MEG/EEG Acquisition
Non-invasive electrophysiology data will be acquired at the Saint Louis University MEG facility. This facility employs a Magnes 3600 (4DNeuroimaging, San Diego, Ca) with 248 magnetometers within a shielded room with 64 EEG Voltage Channels and 23 MEG reference channels (5 gradiometer, and 18 magnetometer). Five current coils attached to the subject, in combination with structural imaging data and head surface tracings, are used to localize the brain in geometric relation to the magnetometers. The same coils are used to monitor and partially correct for head movement during the MEG acquisition session. Auditory (earphone), visual (screen external to shielded room) and somatosensory (air-driven tactile) stimuli can be delivered.
What will MEG/EEG add to the Connectome?
The Connectome Project is largely focused on the communication networks in the brain that are active during quiet rest or when people perform well-specified tasks. Because MEG and EEG measure neuronal activity in “real time” the connections activated either at rest or during task can be measured, giving us a picture of the dynamic interactions among brain networks. MEG has much greater temporal resolution than fMRI so MEG-based analysis provides high temporal resolution data for analyzing the neuromagnetic correlates of fMRI connectivity, its time-frequency content, and temporal interactions. Our preliminary results show good correspondence between fMRI and MEG connectivity patterns in the resting state (see Resting-State MEG). In addition, task-evoked MEG/EEG data will provide rich temporal information that can shed light on information flow among relevant nodes of key cognitive networks. The rich bandwidth of MEG/EEG data and its relationship to synaptic population activity will provide valuable additional information such as cross-frequency interactions and will enable inferences about heritable aspects of MEG patterns.
Figure 3: A subject undergoing an EEG/MEG scan.