- >>
- Healthy Adult Studies
- HCP Young Adult
- Project Protocols Detail
Components of the Human Connectome Project - Pulse Sequences
Two groups of pulse sequences have revolutionized neuroimaging research, diffusion imaging and functional imaging (fMRI). Both rely on image formation via the echo planar imaging (EPI) pulse sequence.
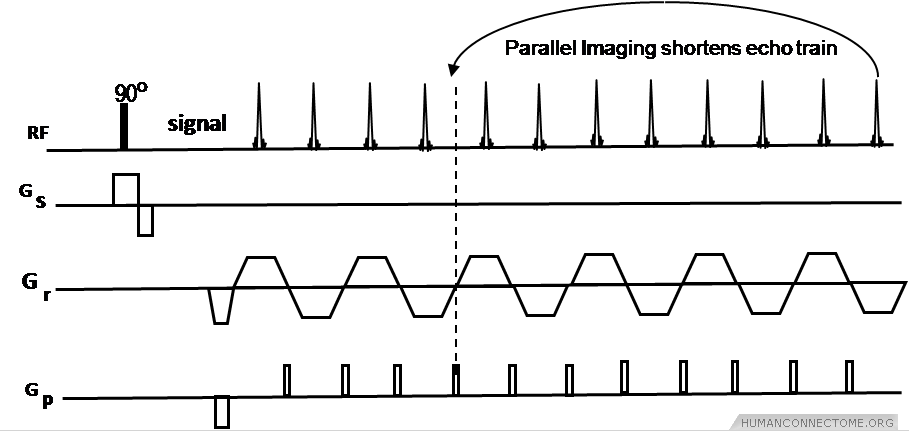
Figure 1: Echo Planar Imaging (EPI) pulse sequence using parallel imaging R-3 reduction or acceleration factor to shorten echo train.
Parallel Imaging EPI for functional MRI.
Parallel imaging with GRAPPA reconstruction (IPAT, Siemens) was evaluated by HCP investigators. As shown in Figure 1, parallel imaging can use 2 to 4 times acceleration factors that in turn reduce echo train length and echo time (TE), improving distortions and, in the case of diffusion imaging, signal-to-noise ratio (SNR) as well. We evaluated all metrics in our comparisons with expected changes in distortions, susceptibility artifact, SNR and scan times. This HCP Phase I work was performed using the new multichannel array coils that were developed on the 3T for best parallel imaging performance, and for increased sensitivity of the coil (approximately a factor of 2 to 3 higher SNR than current 12 channel coils).
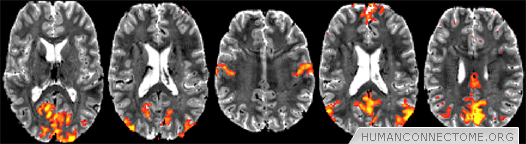
Figure 2: Preliminary resting-fMRI independent component analysis (ICA) results from 7T at 1.5mm isotropic resolution, single-subject data. 50-dimensional ICA shows exquisite cortical detail in several resting networks: low-level visual, higher visual, sensorimotor, primary default mode network (DMN) and secondary DMN.
Faster Whole Brain coverage for fMRI and Diffusion Imaging.
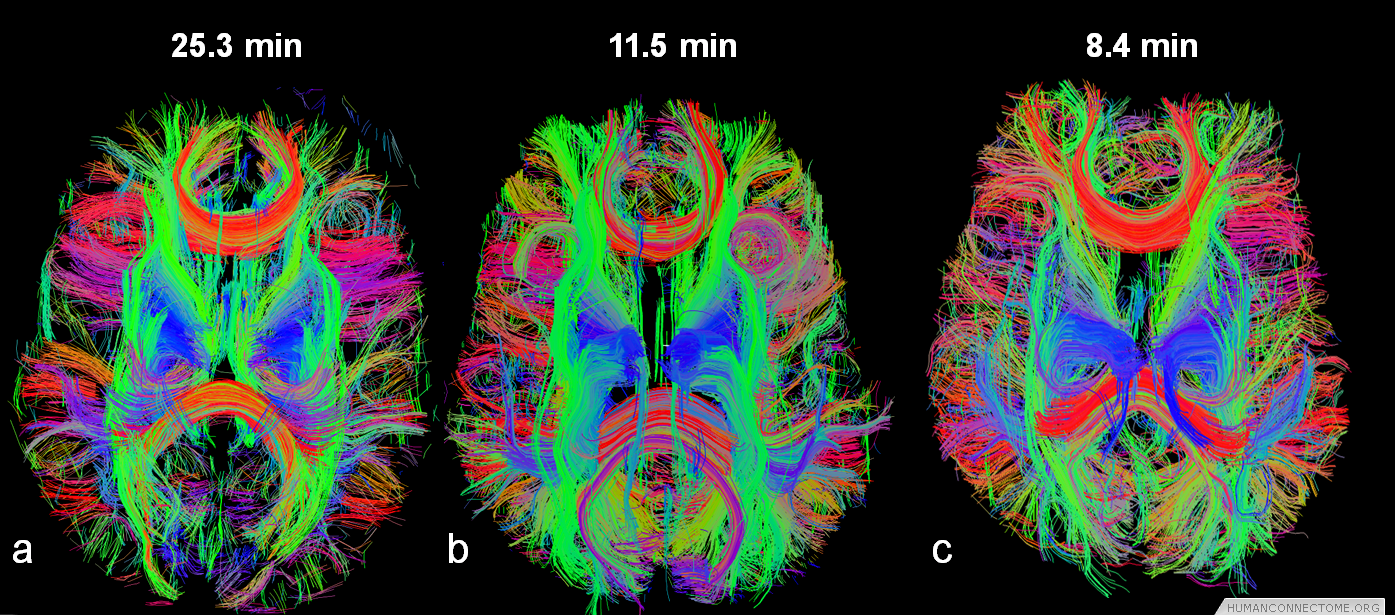
Encoding two or more images in each EPI echo train becomes highly efficient to reduce total scan time. The relatively long encoding time of the HARDI diffusion preparation pulses are applied before the EPI image readout and, therefore, the diffusion pulses can be shared by several images simultaneously. Using 3 images per EPI readout reduces the scan time to by two thirds, from 24 minutes to 8 minutes, and permits HARDI scans to be obtained with higher spatial resolution and/or greater number of b-values in shells or Cartesian coverage of q-space.
We introduced a modification of the multiband approach described by Moleller et al (2010) by combining it with the SIR approach (Feinberg et al 2002, Reese 2009) to achieve multiplicative accelerations (Feinberg et al. 2010). The approach, leading to shorter times for whole brain coverage, was demonstrated to improve statistical significance for the detection of the resting state networks in resting state fMRI and substantially shorted HARDI imaging times. The OT1 team built upon his work to optimize acquisition parameters for data collection using this approach in HCP Phase II.
Application of these Methods to 7T is challanging because at 7T, use of conventional parallel imaging (IPAT in Siemens environment) along the phase encode dimension is mandatory for good image quality. Even though the original slice accelerated multiband imaging in the brain for fMRI (Moeller et al., 2010) was developed for 7T, achieving the same high slice acceleration factors now feasible at 3T requires strategies for improved image reconstruction in the presence of high parallel imaging (IPAT) factors along the phase encode dimension. Accomplishing this for data acquisition at 7T for a subset of HCP participants is a present goal of OT1.
Further Reading:
- Moeller S, Yacoub E, Auerbach E, Strupp JP, Harel N, Ugurbil K. Multi-band Multi-slice GE-EPI at 7 Tesla, with 16 fold acceleration using Partial Parallel Imaging with application to high spatial and temporal whole brain fMRI. Magn Reson Med. 2010, 63(5): 1144-43.
- Feinberg DA, Reese TG, Wedeen VJ. Simultaneous Echo Refocusing in EPI. Magn Reson Med. 2002, 48(1):1-5.
- Reese, T.G., T. Benner, R. Wang, D.A. Feinberg, and V.J. Wedeen, Halving imaging time of whole brain diffusion spectrum imaging and diffusion tractography using simultaneous image refocusing in EPI. J Magn Reson Imaging, 2009, 29(3): p. 517-22.
- Feinberg, D.A.; S. Moeller, S.M. Smith, E. Auerbach, S. Ramanna, M.F. Glasser, K.L. Miller, K. Ugurbil, and E. Yacoub. Multiplexed echo planar imaging for sub-second whole brain fMRI and fast diffusion imaging. PLoS ONE, 2010. 5(12): p. e15710.