- >>
- Healthy Adult Studies
- HCP Young Adult
- Project Protocols Detail
Components of the Human Connectome Project - MR Pre-Processing
Susceptibility artifact correction.Scanned objects have variable magnetic susceptibility, leading to a complex pattern of field inhomogeneities. These cause image distortions and dropout in most diffusion and fMRI acquisitions, with consequences for estimation of connectivity. We have developed new methods for data acquisition and processing that minimize these artifacts and optimize our ability to reduce them further in the data processing.
In the case of diffusion this includes acquisitions that utilize multiple phase-encode directions, moving the artifacts around the image before being recombined. In the case of fMRI, we are utilizing parallel imaging to minimize the distortions, in conjunction with the acquisition of separate fieldmaps to measure the perturbed magnetic field, allowing correction at the data processing stage.
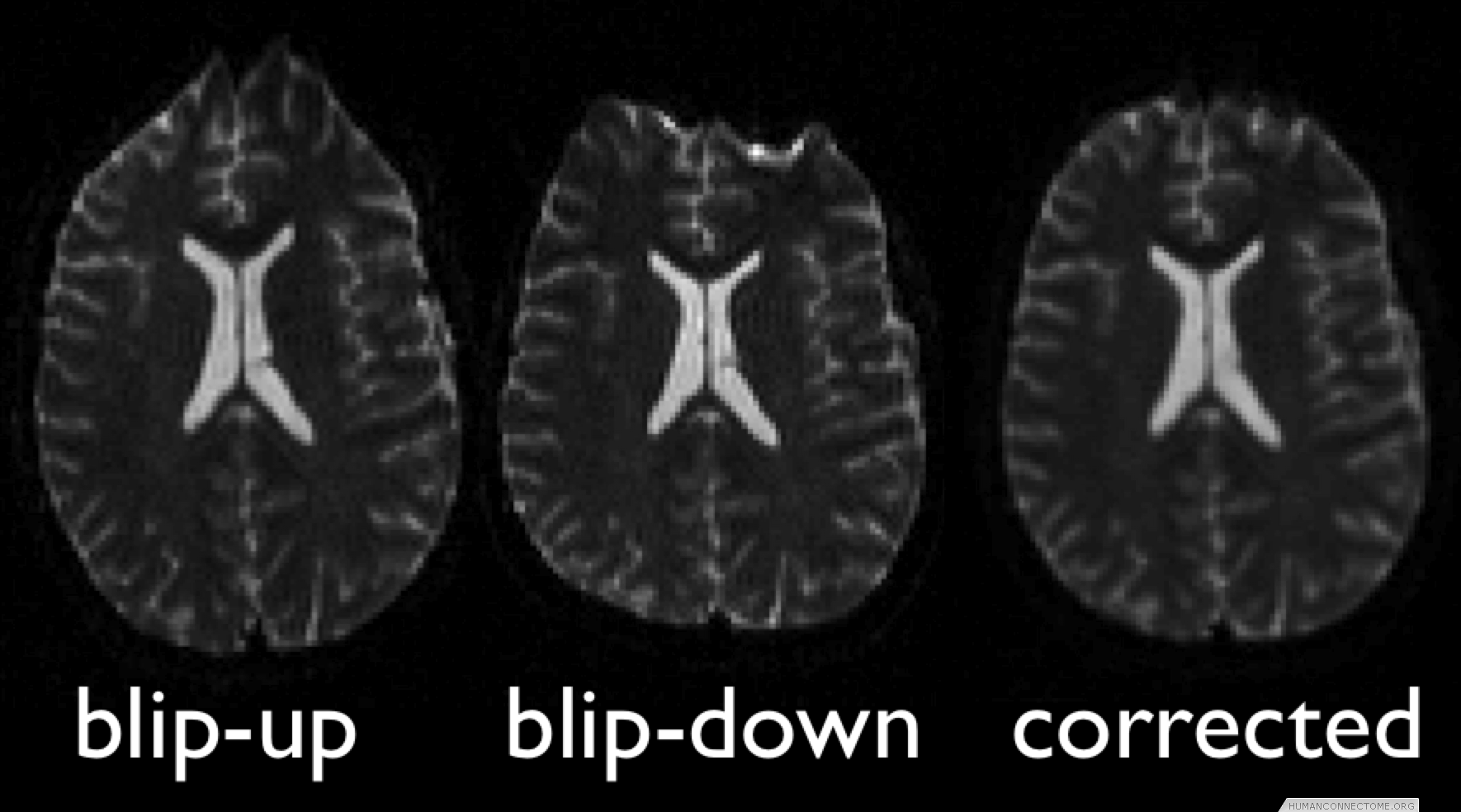
Figure 1: Example 3T diffusion data collected with two phase encoding directions, combined to give undistorted data.
Eddy-current and head-motion correction. Strong and rapidly switched diffusion gradients cause scaling and shear in the phase-encode direction, and translations that may vary with slice position. Standard approaches for correction can be inaccurate given the low SNR of raw diffusion data, particularly at high b-values. We have developed new methods to ameliorate this by estimating eddy current and head motion effects simultaneously with prediction of the data for each of the different diffusion directions.
Other MRI and physiological artifacts. Other artifacts such as RF field inhomogeneities, gradient nonlinearities, EPI ghosting, spiking and chemical shift are being minimized during acquisition using standard techniques and further reduced where necessary in data processing. Residual ghosting and spiking in fMRI are being identified and removed using methods such as independent component analysis. Physiological effects such as cardiac pulsation, breathing-related artifacts and residual head motion effects are being minimized through a combination of model-based and data-driven methods.
Quality control. In addition to specific methods for artifact removal, we are implementing a fully automated analysis pipeline for general QC, which are being executed by ConnectomeDB immediately after data upload. We are utilizing low-level tools that are integrated into our automated pipeline, such as linear and nonlinear registration, brain-extraction, and tissue-type segmentation, to automatically detect/correct problematic data. We are generating summary statistics on both signal-to-noise ratio and contrast-to-noise ratio, and comparing with established norms, to provide immediate warning of hardware problems. We are also generating QC tests that will be applied to data further along the processing pipeline, to give further indications of outlier data sets.
Anatomically constrained analysis and visualization. We are using segmentations and cortical surface models derived from high quality T1-weighted data to underlie much of the analysis and visualization of all modalities. We have established a pipeline that begins with a T1 image and ends with cortical surface models and subcortical segmentation. We are therefore able to combine data across subjects both in 3D standard space and on 2D ‘standard-mesh’ surfaces.
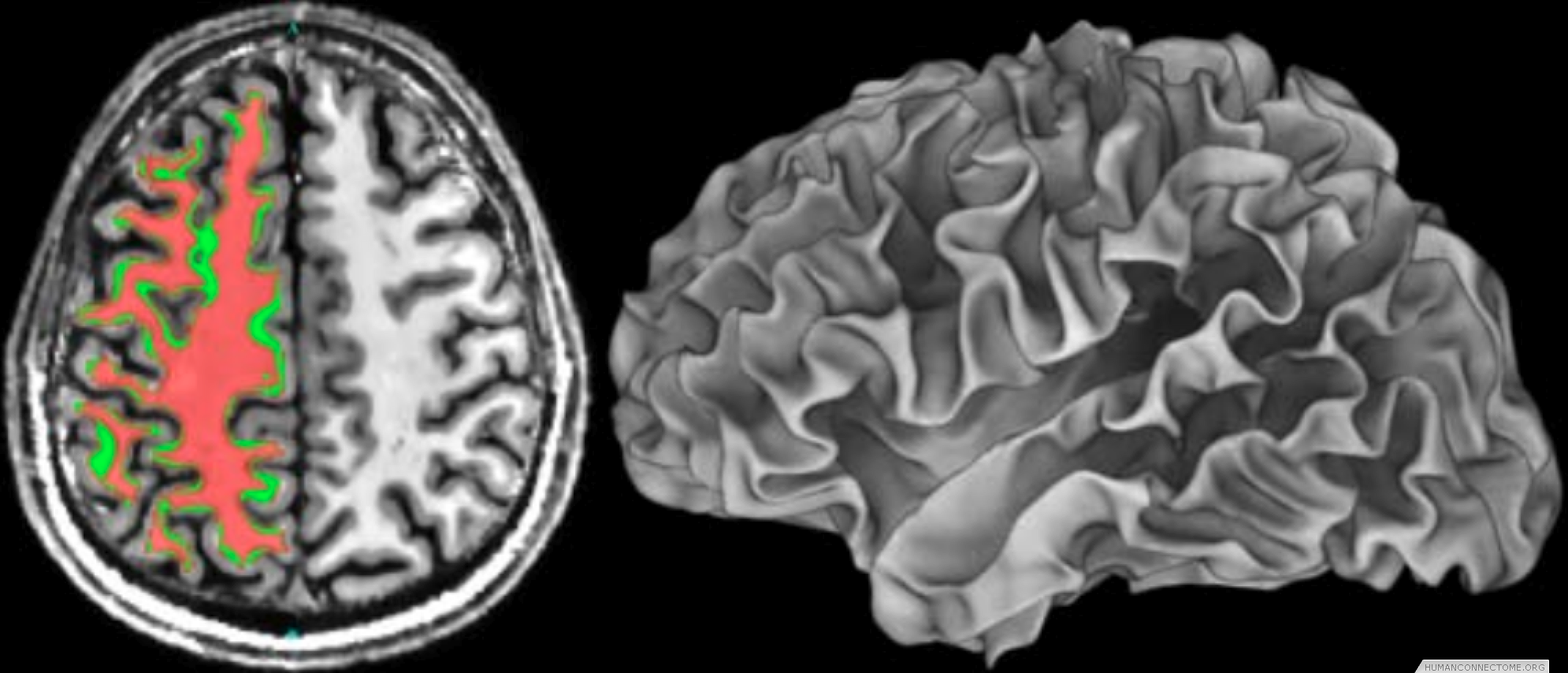
Figure 2: Left: Example T1-weighted image acquired at 7T with white matter segmentation overlaid. Right: White matter surface rendering.